Curing Death
Approximating biology in silicon will allow us to understand senescence, error-correct cellular information loss, and cure death.
Side note: this, along with other posts here, is constantly updating as I learn more and my thoughts develop.
Recently I read the great article can a biologist fix a radio? I recommend anyone interested in biology but not currently working in it give it a read. It really helps someone like me, used to the clean perfection of abstracted bit processing, to grasp just how difficult a problem the entire medical industry is dealing with, and the flaws in the current reductionist / empiricist approach. It's the hardest debugging problem of all time.
By far the worst part of this problem is just how difficult it is to run tests. When I face a problem with my code, I can run it 50 different ways in a few minutes, testing out different inputs to see how it behaves. Now imagine how long it would take to find a bug if you could only run your code a few times over 3 years, and each time you hit run it costs $41,000. Quite the challenge!
And yet that's the exact situation researchers are contending with. The costs of clinical trials have spiraled out of control, and FDA approval is harder than ever to recieve. So how does this get solved? The obvious answer is political. The FDA should be slimmed down, the approval process made more transparent, the discovery process subsidized. But I'm not interested in political solutions.
Taking inspiration from aerospace
The aerospace industry faced a very similar problem in the mid-20th century. As aircraft got bigger, wind tunnels needed to grow to fit models that would have similar behavior as the full scale version. As aircraft got faster, wind tunnels needed to get more powerful, eventually requiring supersonic wind tunnels to model transsonic and supersonic flight charactaristics. These tunnels are incredibly expensive, and still can only operate on scale models.
Modern aircraft designs spend far less time in the tunnel because most design and testing happens ahead of time on comparatively cheap computational fluid dynamics (CFD) simulations. These sims are able to model air flow over a rigidbody, predict drag, lift, and structural stresses on the airframe, all before any physical model needs to be constructed.
Today an aerospace engineer can design an airframe entirely on a computer, and have reasonable confidence it will work well when a scale model is created. How can we acheive the same thing in biology?
First steps
Computational biology isn't new. In the 1990's, the Human Genome Project sought to map out over 85% of the human genome, which it achieved in 2003. Today they've completed over 99.7% of the genome, declaring a "complete genome" has been mapped.
Since then, the price of mapping a genome has seen faster progress than Moore's Law.
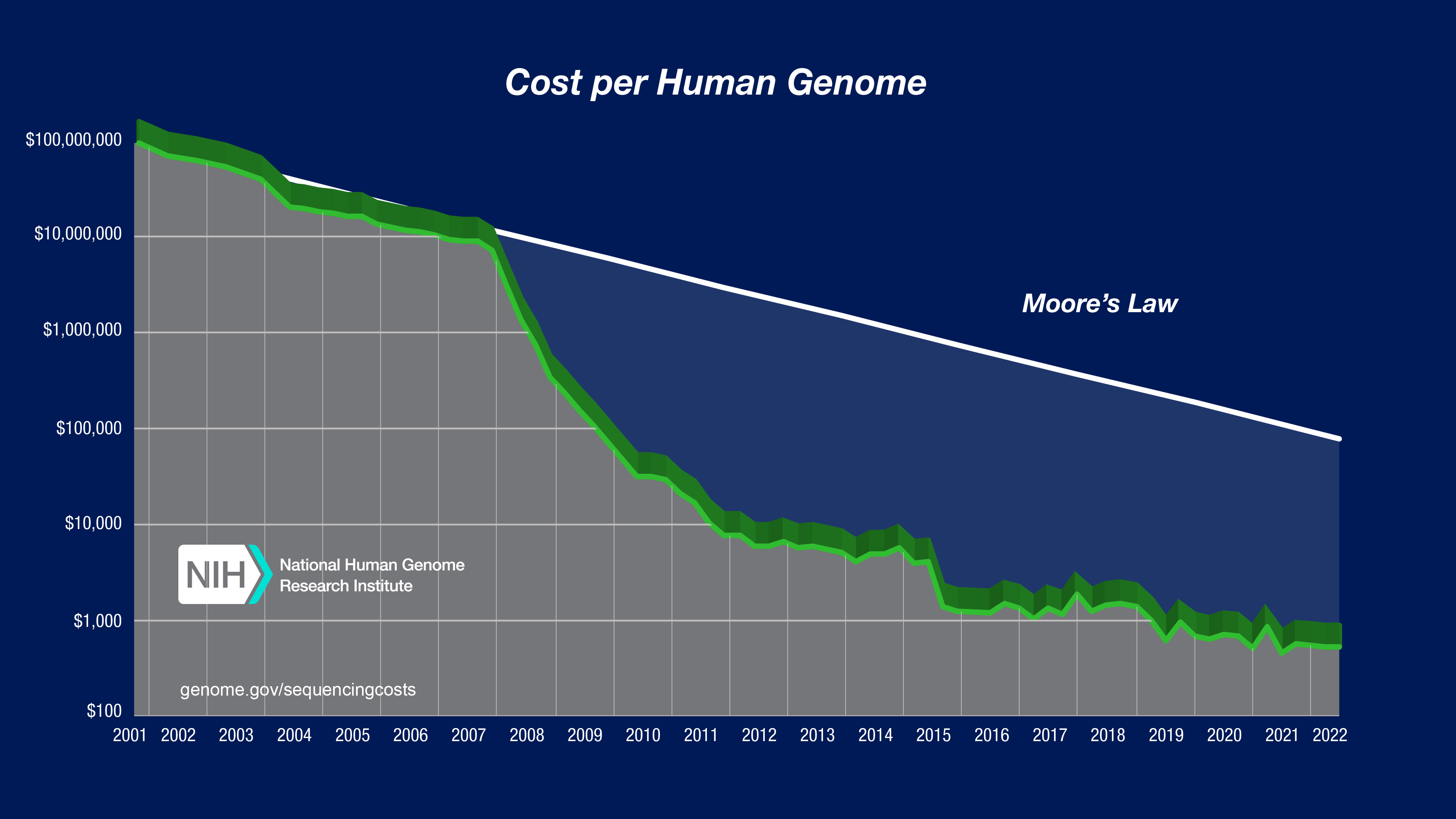
With next-generation sequencers and much faster computers, you can purchase whole genome sequenceing for $300-$900.
Advances in genome mapping allowed computers to start processing and learning from genome sequences. Given the billions of nucleotides in a genome, how can we know what each of them does? This question is vital to understanding genetic diseases, personalizing medicine, predicting future issues, and understanding why certian people's bodies act differently than others.
DNA to Proteins
So we can sequence a genome. What's next?
We have a huge sequence of nucleotides, but what do they do in the body? One of the main functions DNA goes through is a pipeline of processes known as transcription, splicing and translation that turn DNA into proteins.
Transcription
DNA is copied to RNA in a process called transcription. What gets transcribed is determined by transcription factors, proteins that bind to parts of the DNA and control transcription. Transcription is also impacted by promoters and enhancers, segments of DNA outside of the part being transcribed that have binding sites for transcription factors and use them to regulate transcription. Promoters are directly upstream from the gene, while enhancers can be distant.
Splicing
Once the initial RNA strand is created through transcription, segments of it are cut out. The parts cut out are called introns, and the parts kept are called exons. Once the introns are cut out, the exons are glued together in-order. The splicing process can allow one transcribed gene to be turned into many different final RNA strands. The 20,000 genes present in the human genome can be turned into over 70,000 RNA strands.
Translation
Once the splicing is complete, and the exons are glued back together, the RNA strand is sent to the ribosome to construct the protein. The ribosome reads in the strand in groups of 3 nucleotides, called codons, which directly map to 20 different amino acids. These amino acids are strung together to form the unfolded protein, which then folds into it's final functional form.
At the end of this process we're left with protiens, one of the most important classes of molecules in the body. It's vital we can model these if we're to understand how the body works on a cellular level. For more detail on this process, here's a great post.
AlphaFold
Protein behavior is determined by their shape, and after they are constructed from DNA, they go through a complex process of folding to achieve their 3D form.
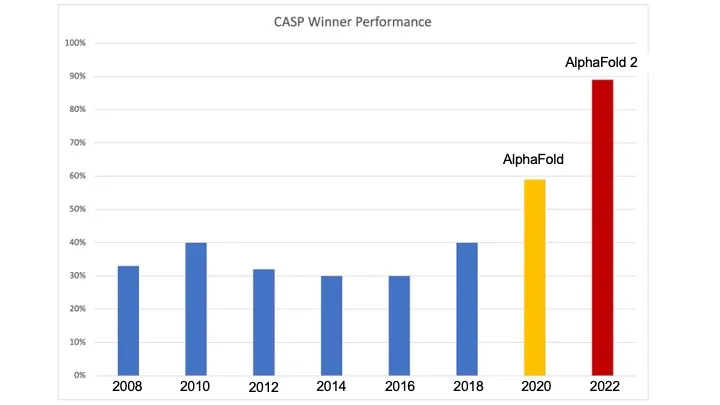
The CASP competition measures teams ability to predict the 3D structure of proteins after they fold, given the protein's amino acid makeup. The competition has taken place every 2 years since 1994, and each year has seen roughly stable performance from the competitors. Until 2018.
Deepmind had been using deep learning to acheive incredible feats, like defeating the world champion of Go and then producing models with even less pre-programmed rules that performed even better. Neither Deepmind nor anyone else know the limits of what these techniques can learn, but clearly they are formidable approximators.
Since the process of protein folding is a function which we have many inputs and outputs of, can deep learning approximate this process as well? In 2018, Deepmind clearly answered yes by trouncing all competitors and producing vastly superior predictions. They followed up the performance with a larger and more powerful model in 2020, producing predictions accurate enough to be useful to the medical industry.

Clearly deep learning can approximate complex biological processes we have little understanding of. So how far can we take this principle? Can any biological process be approximated, simulated, and predicted, all the way up to a full human body?
The data problem
Yes and no. We've seen that powerful deep learning models posess formidable modeling capacity. But they also require large amounts of data to learn from. In the protein folding problem, AlphaFold 1, 2 and 3 all had access to over 100,000 experimentally validated examples of protein structures.
We don't have access to data of that magnitude for most biological processes. So how can we get it? Do we even need it?
Let's take a look at some processes we may want to model next, and the pathway to acheiveing full-cell simulation, often regarded as the holy grail of computational biology.